Page Not Found
Page not found. Your pixels are in another canvas. Read more
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas. Read more
About me Read more
This is a page not in th emain menu Read more
Published:
Useful resource for every coding enthusiast
Published:
Useful resource for every coding enthusiast
Basics of GNNs with useful resources to start with graph neural networks Read more
A compilation of the most used commands in bash Read more
Online summer school on machine learning and data sciences that we organized. Read more
Some of the common resources we use in image registration Read more
Some useful CS topics that are not taught as part of the university curriculum. The standard CS curriculum is missing some of these critical topics about the computing ecosystem that could make students lives significantly easier. Read more
A review on Graph transformers Read more
Published in Arxiv, 2019
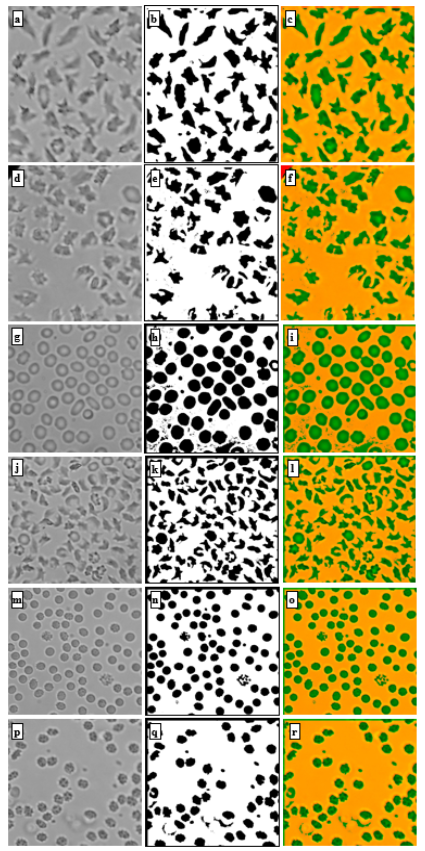
This paper introduces a machine learning-based method for fully automated diagnosis of sickle cell disease of poor-quality unstained images of a mobile microscope. Read more
Published in ESMO MAP, 2021
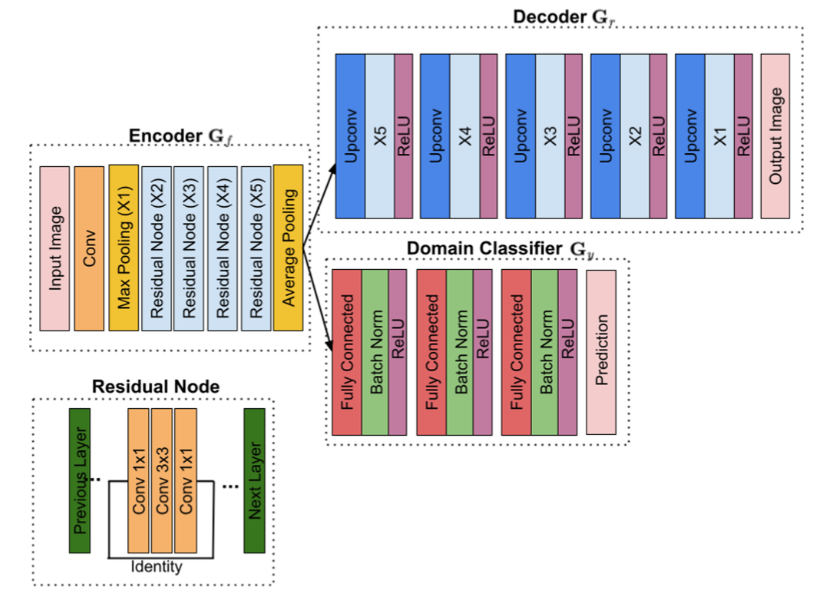
This paper proposes a pre-processing autoencoder that is trained adversarially to the sources of domain variations for mitotic figures detection in histological tumor images. Read more
Published in 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2022
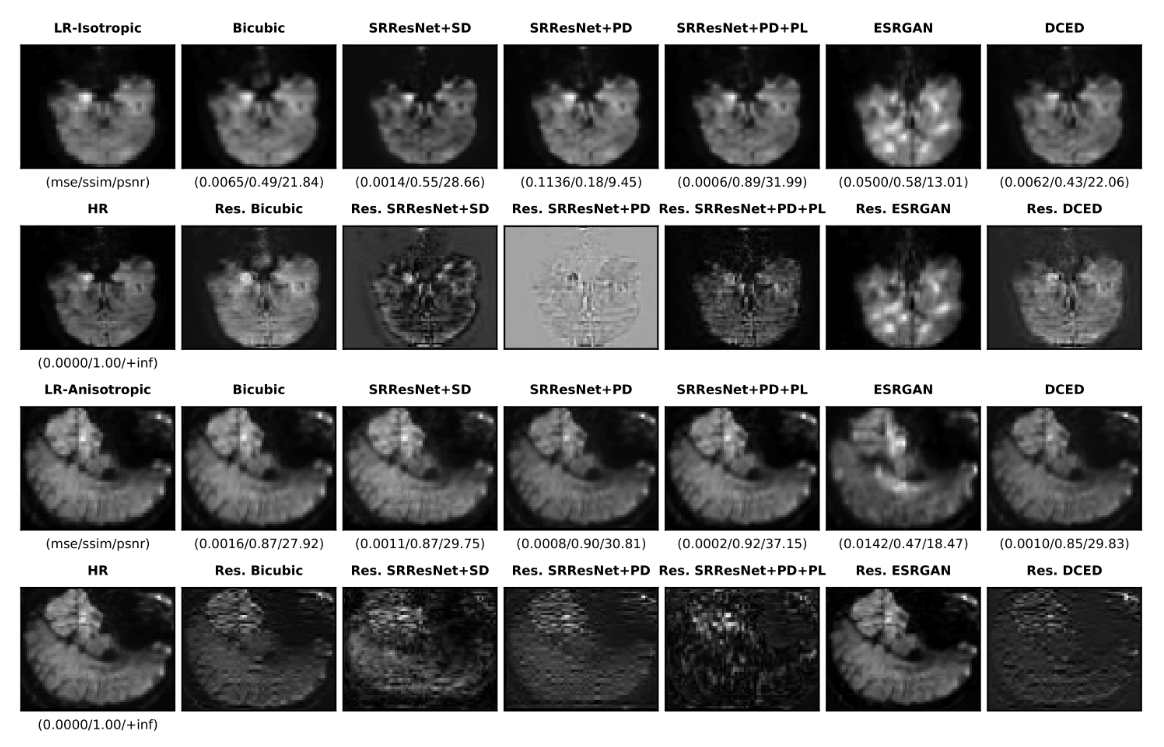
In this paper, we present a super-resolution SR technique for MR images that is based on generative adversarial networks (GANs), which have proven to be quite useful in producing sharp-looking details in SR. Read more
Published in IEEE International Symposium on Biomedical Imaging (ISBI) 2022, 2022
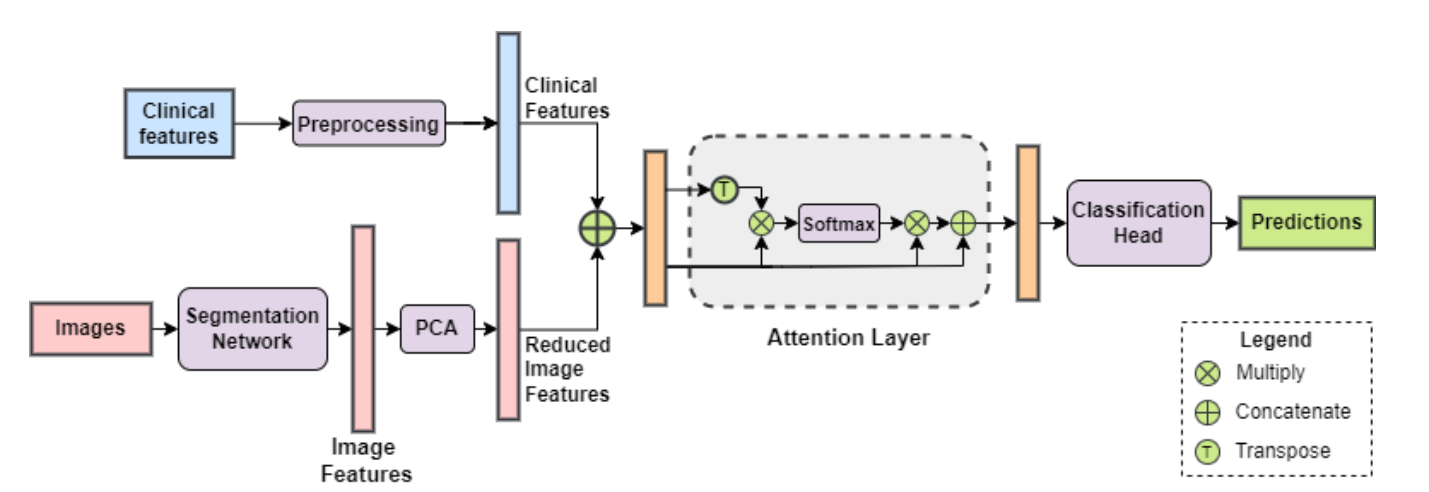
In this paper we propose an attention-based deep learning framework that automatically analyzes renal tumors by fusing both the clinical and imaging features for a more accurate classification of kidney abnormalities. Read more
Published in 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023), 2023
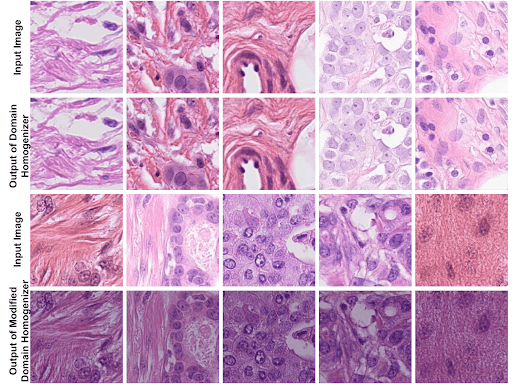
This paper proposes a domain homogenizer for mitosis detection that attempts to alleviate domain differences in histology images via adversarial reconstruction of input images Read more
Published in The 16th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2023, 2023
This paper introduces robust sample specific loss functions for histology image analysis. Read more
Published in IEEE Transactions on Biomedical and Health Informatics, 2023
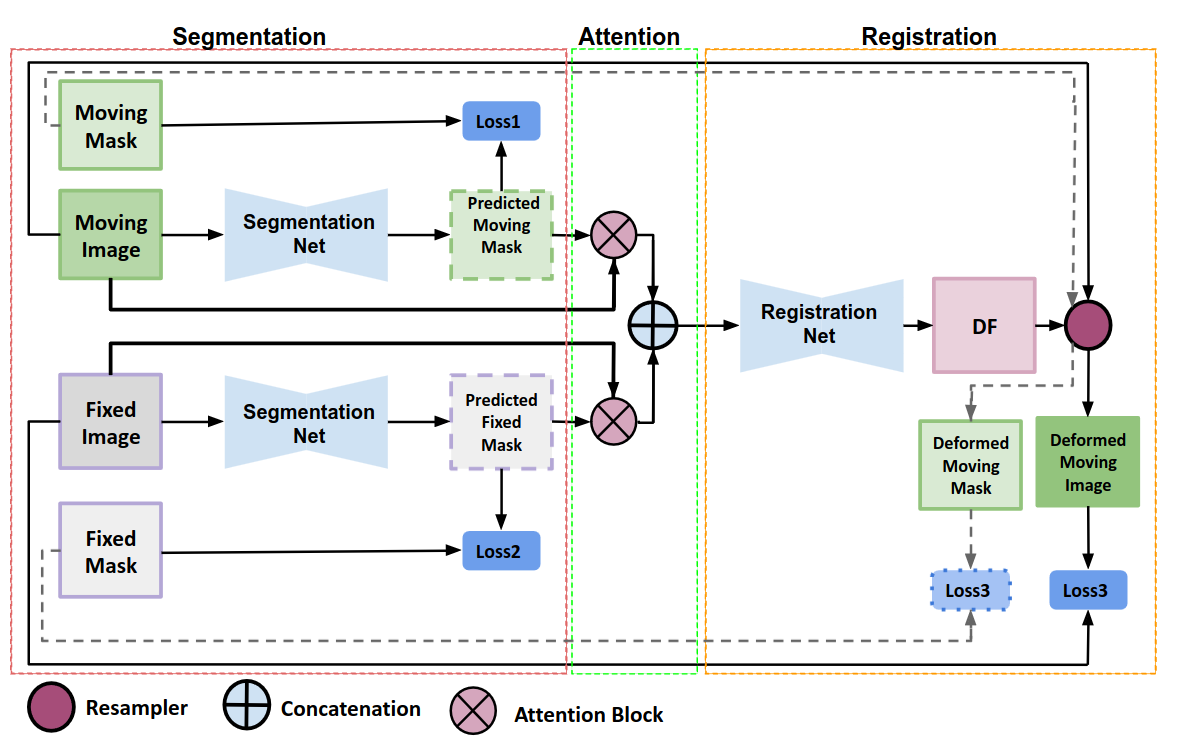
This paper investigates the impact of segmentation onmedical image registration. Read more
Published in WACV2023, 2023
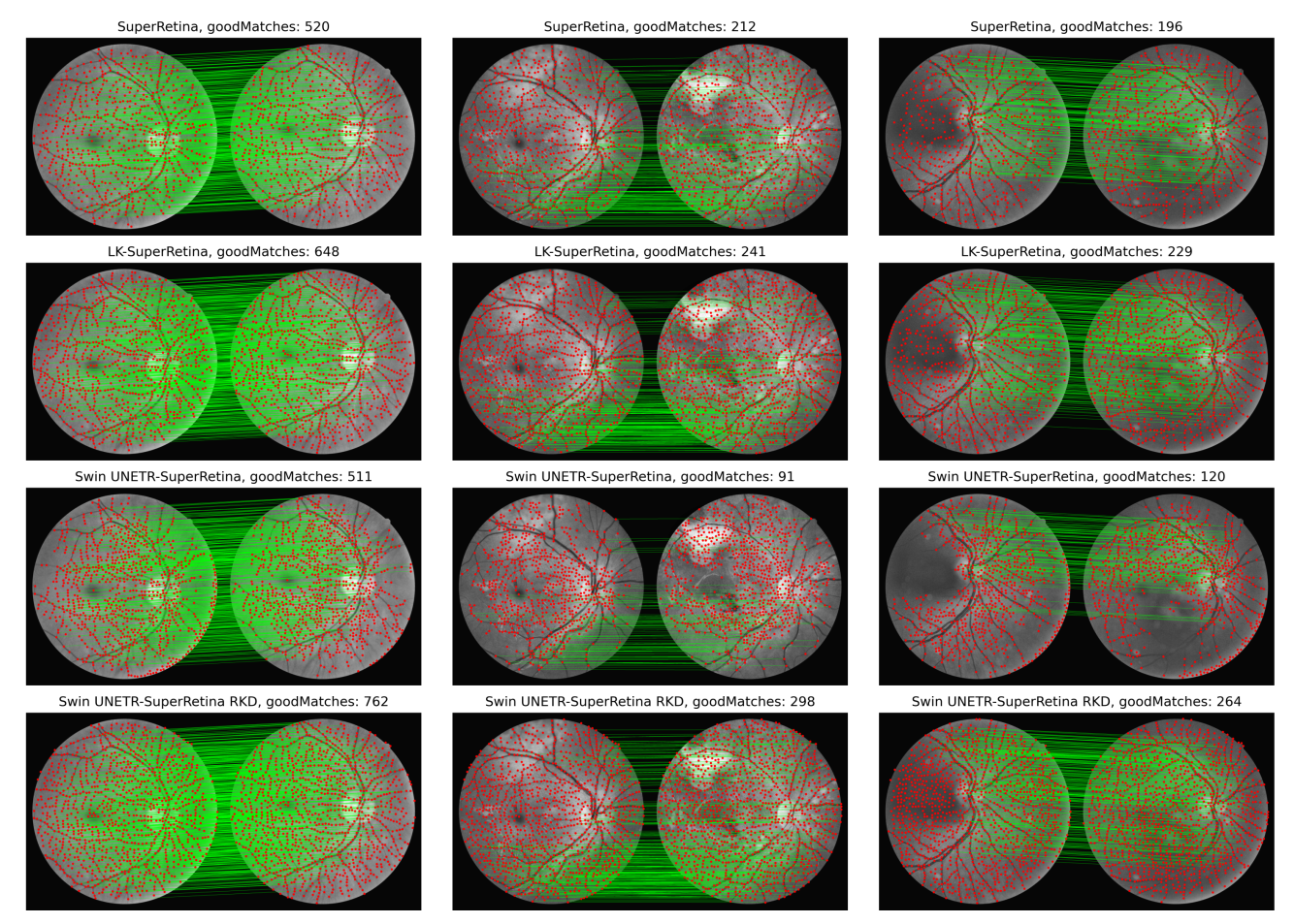
We propose a novel approach based on reverse knowledge distillation to train large models with limited data while preventing overfitting for retinal image registration. Read more
Published in JCO Oncology Practice, 2025
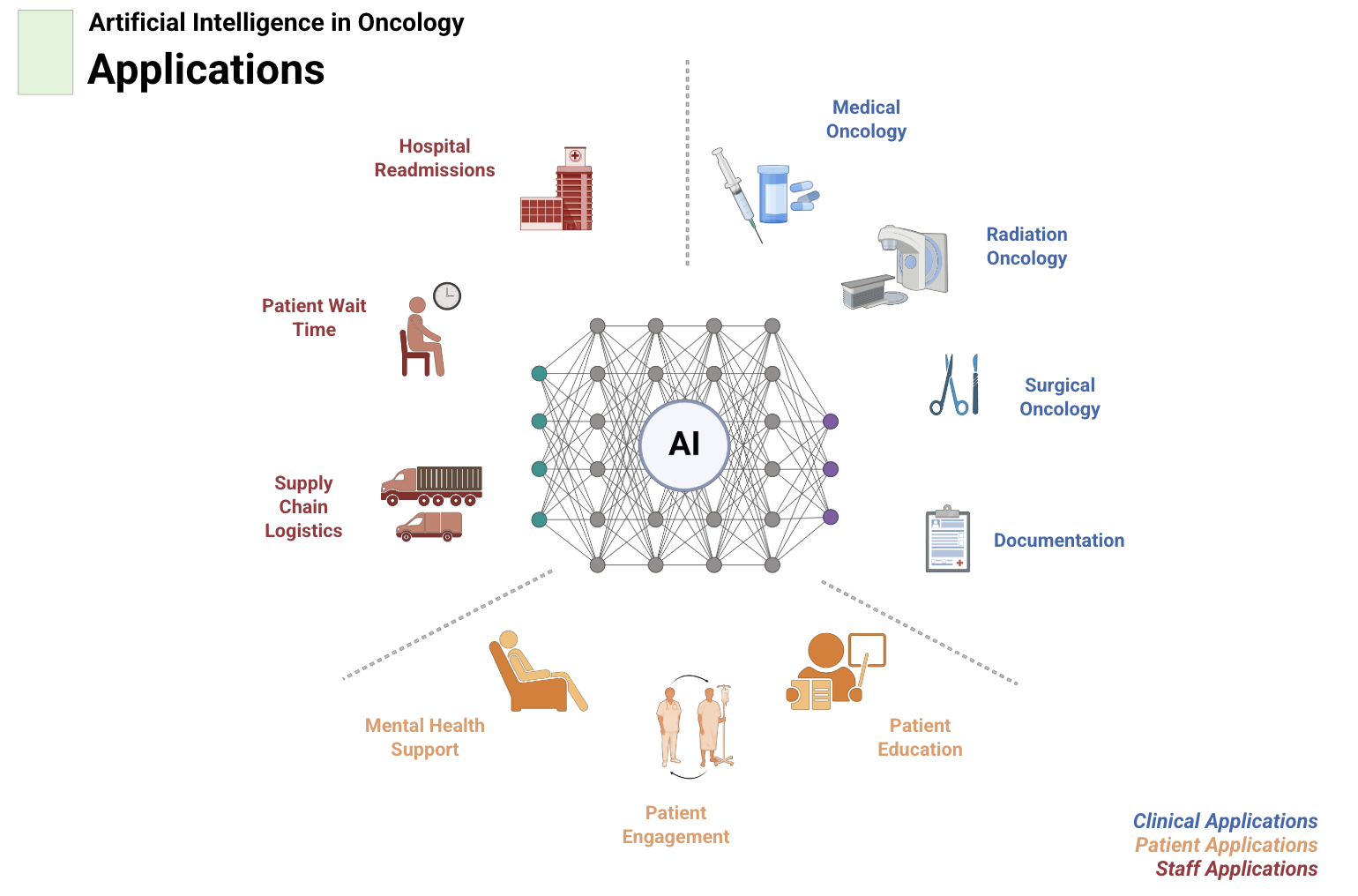
This perspective highlights the opportunities and unmet needs for artificial intelligence in oncology, presented from both the clinician and patient viewpoints, and outlines pathways for equitable and explainable AI adoption in cancer care. Read more
Published in ASCO Conference, 2025, 2025
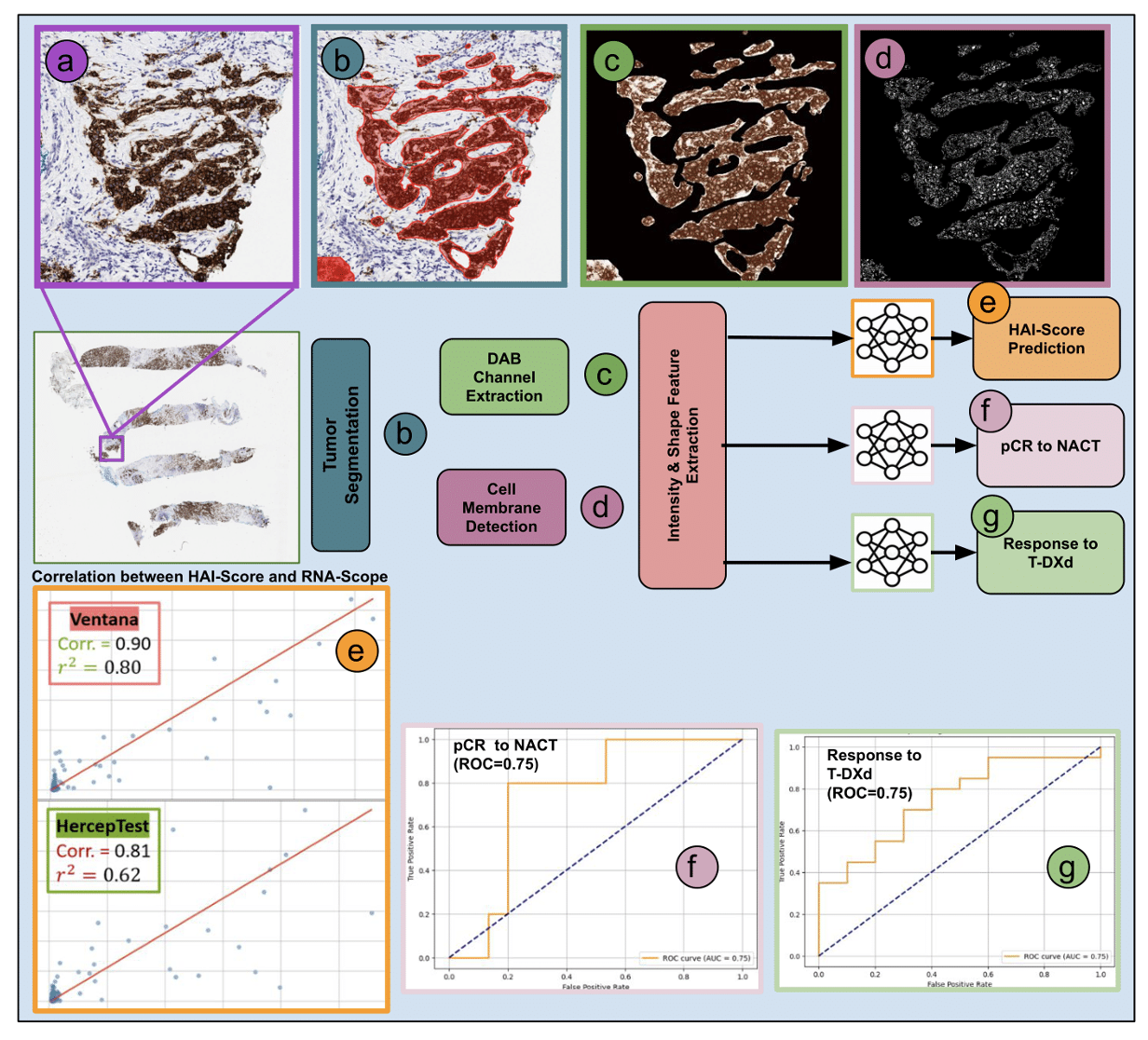
This paper introduces HAI-score, a robust AI-driven approach for quantifying HER2 H-scores from IHC-stained breast cancer samples, providing a reliable alternative to manual scoring and FDA-approved assays. Read more
Published:
 Qualcomm Innovation Fellowship winners announced - Interact with Secretary of MEITY and Principal Scientific Advisor Abstract:
Qualcomm Innovation Fellowship winners announced - Interact with Secretary of MEITY and Principal Scientific Advisor Abstract:
Prostate cancer is the second largest cause of cancer deaths in men worldwide. The prostate is a dense organ that is surrounded by many other organs. Consequently, it is difficult to locate suspicious regions within the prostate for a diagnostic biopsy whose needle cannot be guided easily using real-time and portable imaging techniques, such as ultrasound (US) imaging. The standard diagnostic procedure is random needle biopsy, in which there is always a chance to miss a small tumor mass of high grade. On the other hand, the tumor contours are better visible in magnetic resonance imaging (MRI). Therefore, a fusion of the two imaging modalities has the potential to combine their advantages. However, the existing solutions require manual matching of fiducial points on the two modalities, and they do not simulate real-time organ displacement and deformation during biopsy or surgery. Furthermore, the potential of the US imaging has not been exploited very well till now. For example, the wave interference pattern in the US, which is considered as “speckle noise,” contains a lot of information about the structure and pathobiology of the organ. But, this information is lost and left unused when the US images are despeckled. Instead, the information in the wave interference pattern may be usable, if disentangled. Read more
Published:
 Conference proceedings on our paper: Perceptual CGAN for MRI Super-resolution Abstract:
Conference proceedings on our paper: Perceptual CGAN for MRI Super-resolution Abstract:
Capturing high-resolution magnetic resonance (MR) images is a time consuming process, which makes it unsuitable for medical emergencies and pediatric patients. Low-resolution MR imaging, by contrast, is faster than its high-resolution counterpart, but it compromises on fine details necessary for a more precise diagnosis. Super-resolution (SR), when applied to low-resolution MR images, can help increase their utility by synthetically generating high-resolution images with little additional time. In this paper, we present an SR technique for MR images that is based on generative adversarial networks (GANs), which have proven to be quite useful in producing sharp-looking details in SR. We introduce a conditional GAN with perceptual loss, which is conditioned upon the input low-resolution image, which improves the performance for isotropic and anisotropic MRI super-resolution. Read more
Published:
 Conference proceedings on our paper: “WSSAMNet: Weakly Supervised Semantic Attentive Medical Image Registration Network” Abstract:
Conference proceedings on our paper: “WSSAMNet: Weakly Supervised Semantic Attentive Medical Image Registration Network” Abstract:
We present WSSAMNet, a weakly supervised method for medical image registration. Ours is a two step method, with the first step being the computation of segmentation masks of the fixed and moving volumes. These masks are then used to attend to the input volume, which are then provided as inputs to a registration network in the second step. The registration network computes the deformation field to perform the alignment between the fixed and the moving volumes. We study the effectiveness of our technique on the BraTSReg challenge data against ANTs and VoxelMorph, where we demonstrate that our method performs competitively. Read more
Published:
 Abstract:
Abstract:
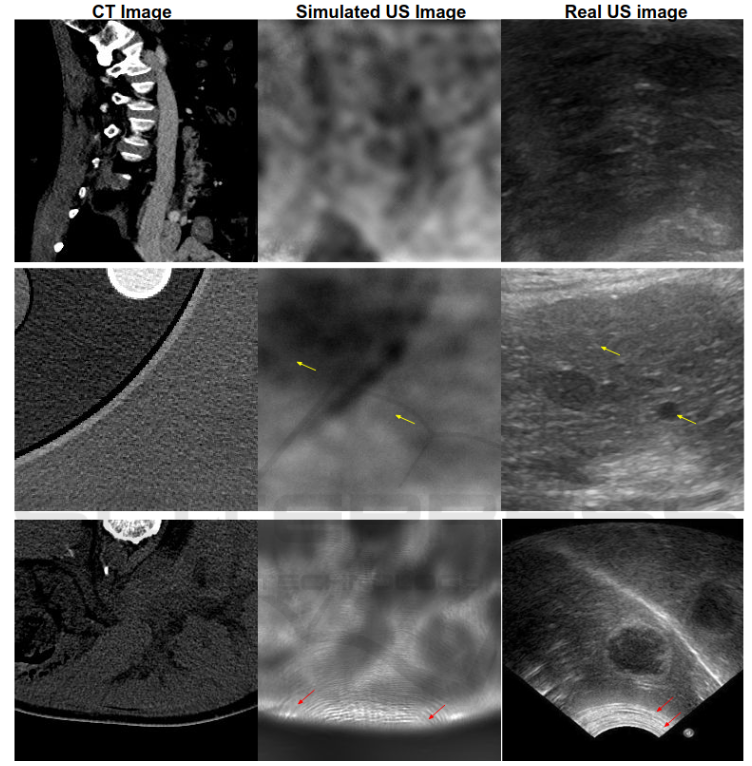
Anatomical information in ultrasound (US) imaging has not been exploited fully because its wave interference pattern (WIP) has been viewed as speckle noise. We tested the idea that more information can be retrieved by disentangling the WIP rather than discarding it as noise. We numerically solved the forward model of generating US images from computed tomography (CT) images by solving wave-equations using the Stride library. By doing so, we have paved the way for using deep neural networks to be trained on the data generated by the forward model to simulate the solution of the inverse problem, which is generating the CT-style and CT-quality images from a real US image. We demonstrate qualitative features of the generated images that are rich in anatomical details and realism. Read more
Published:
 Conference proceedings on our paper: Improving Mitosis Detection Via UNet-based Adversarial Domain Homogenizer Abstract:
Conference proceedings on our paper: Improving Mitosis Detection Via UNet-based Adversarial Domain Homogenizer Abstract:
The effective counting of mitotic figures in cancer pathology specimen is a critical task for deciding tumor grade and prognosis. Automated mitosis detection through deep learning-based image analysis often fails on unseen patient data due to domain shifts in the form of changes in stain appearance, pixel noise, tissue quality, and magnification. This paper proposes a domain homogenizer for mitosis detection that attempts to alleviate domain differences in histology images via adversarial reconstruction of input images. The proposed homogenizer is based on a U-Net architecture and can effectively reduce domain differences commonly seen with histology imaging data. We demonstrate our domain homogenizer’s effectiveness by showing a reduction in domain differences between the preprocessed images. Using this homogenizer with a RetinaNet object detector, we were able to outperform the baselines of the 2021 MIDOG challenge in terms of average precision of the detected mitotic figures. Read more
Published:
 I presented our paper “Reverse Knowledge Distillation: Training a Large Model Using a Small One for Retinal Image Matching on Limited Data” at WACV 2024.
I presented our paper “Reverse Knowledge Distillation: Training a Large Model Using a Small One for Retinal Image Matching on Limited Data” at WACV 2024.
This work achieved state-of-the-art performance for retinal image registration by leveraging knowledge transfer from a small to a large model.
Published:
 I was invited to give a TEDx Salon Talk on how AI tools are improving equitable breast cancer diagnosis and treatment.
I was invited to give a TEDx Salon Talk on how AI tools are improving equitable breast cancer diagnosis and treatment.
In this talk, I shared my research journey, highlighting how computational pathology and explainable AI can bring precision diagnostics to underserved populations.
Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.